Courses
STEM Summer Research Dublin Courses
You will earn 6 research credits over 8 weeks, conducting faculty-supervised, hands-on, directed study research projects with results that will culminate in the preparation of a research paper. You will complete a minimum of 240 hours on research in and out of the laboratory.
Faculty mentors will work closely with you to direct your continued growth and knowledge development in the chosen research topic discipline.
- Please review your project with your academic or study abroad advisor to ensure it will transfer back to your home school and that you are following your home school’s policies.
Choosing Your Research Project
- Review Project titles and descriptions below.
- List 3 (in order of preference) in your Academic Preferences Form, using DUBL as the course code.
- Program is highly individualized, with limited enrollment.
- You will need to complete a brief Literature Review in consultation with your research supervisor prior to departure before the start of the program. More details here.
- We encourage you to contact Arcadia Abroad Academic Advisors, Leah Cieniawa or Richard Evans II, to discuss your particular research interests further.
- Additional projects for Summer 2026 may be added as they become available.
| Course ID | Title | Credits | Syllabus |
|---|---|---|---|
| DUBI RSLW 392S | International Independent Research in STEM Fields | 6 |
Summer 2026 Research Projects
Investigating the tole of disease-causing proteins in motor neurons
Supervisor: Dr niamh O'sullivan
My lab studies inherited forms of motor neuron disease, particularly hereditary spastic paraplegia (HSP). Individuals with HSP develop weakness in their legs leading to difficulties walking which is caused by degeneration of the very longest motor neurons. Extensive work in recent years has successfully identified many of the genetic causes underpinning HSP, but there are currently no treatments to prevent, cure or even to slow the course of these diseases. To address this, my lab use cutting-edge genetic engineering to generate novel animal models of HSP in the fruit fly, in which we can study the molecular events underpinning this disorder. Recently, researchers in my lab have found that HSP-causing genes play a role in the organisation of the endoplasmic reticulum (ER) network within motor neurons. The aim of your project will be to study how this impaired ER network contributes to neurodegeneration in motor neurons. You will learn various techniques associated with molecular genetics, confocal microscopic image analysis and the assay of behavioural readouts.
Lab website: fniamhy.wixsite.com/osullivanlab
Relevant majors: Biomedical majors
study of the cross-talk between srf and ar in prostate cancer
Supervisor: Dr maria precipe
Background: Current treatments for prostate cancer mainly target the Androgen Receptor (AR), however despite initial response these treatments fail. Serum response factor (SRF) was previously identified as an important transcription factor in in vitro models of castrate-resistant prostate cancer (CRPC) and a cross-talk between AR and SRF was demonstrated.
To further understand this cross-talk, we used mass spectrometry to identify common interactors between these two proteins. The aim of this project is to manipulate the key common interactors to assess cellular response and to study their signaling pathway in prostate cancer cell lines.
Objectives:
- Manipulate common interactors of SRF and AR including HSP70, HSP90, PI3K and Akt using small-molecule inhibitors in a panel of prostate cancer cell lines.
- Assess cell proliferation in response to these inhibitors singly and in combination with enzalutamide (current treatment for CRPC).
- Assess protein expression and activity of several proteins in the pathway after treatments.
Techniques: Cell culture, MTT assays (cell viability), colony forming assay (cell proliferation), Incucyte (cell proliferation for combination treatments), treatment with small molecule inhibitors, western blotting (protein expression) and luciferase assays (protein activity).
Relevant majors: Pharmacology, Genetics, Biochemistry, Molecular Biology
application of cinematic videography for scientific communication
Supervisor: Dr fun man fung
Scientific research is a major global investment and changes people’s lives. In 2024, a projected $2.53 trillion is being allocated worldwide to research and development (R&D). This substantial figure demonstrates the increasing recognition of the importance of scientific innovation in addressing global challenges and driving economic growth. Scientific research is funded by taxpayer’s money. However, science is deemed difficult to comprehend by the public as most of them are not scientists by training. Therefore, there is a need to leverage the technology of videography to bridge the gap between scientists and the public, making scientific research more accessible and engaging. This study aims to provide empirical evidence for the benefits of using cinematic videography as an accessible tool for scientific communication, potentially contributing to a more informed and scientifically literate society.
Researchers on this project will create and employ cinematic videography as the technique to conduct the research. The person will be assigned to combine visually engaging imagery with clear and concise explanations in order to captivate audiences and facilitate a deeper understanding of complex scientific processes. This work is important and could lead to the next stage of a longitudinal research – Using a mixed-methods approach to investigate the effectiveness of cinematic videography.
Relevant majors: Chemistry, Biology, Physics, Geology, Life Sciences
sustainable continuous chemical synthesis of bioactives inspired by nature
Supervisor: PI: associate professor marcus baumann
We wish to offer this unique summer research project that will provide a talented student with the opportunity to acquire and master important research skills in a project at the interface of organic chemistry, sustainability, biology and chemical engineering.
The synthesis and spectroscopic characterisation of a small collection of drug-like building blocks will be targeted with a specific emphasis on exploiting light as a tuneable and sustainable energy source. This will involve bespoke continuous flow reactors equipped with efficient light-emitting diodes (LEDs) available in our lab. Light in the UV-A and visible range of the spectrum will be used to functionalise the molecules studied which will contribute to greener chemical reactions. Continuous flow chemistry (A field guide to flow chemistry for synthetic organic chemists - Chemical Science (RSC Publishing)) is a novel and exciting addition to the chemist’s toolbox that allows to perform chemistry in a safer, more effective and sustainable manner yielding readily scaled and automated processes that are highly desirable in academia and industry alike. Flow chemistry has been identified as one of the 10 emerging technologies that will change the world: IUPAC announces the top ten emerging technologies in chemistry - IUPAC | International Union of Pure and Applied Chemistry. This approach will avoid the isolation of unstable molecules and provides a powerful and streamlined route into important bioactive structures that will be studied by biologists and medicinal chemists. The ability to automate such processes and couple them with AI and machine learning is highly advantageous as it circumvents tedious downstream processing to directly give clean products.
The successful student will be embedded in our international research group (currently 10 PhDs, 2 postdocs) and gain new skills in chemical synthesis, purification, spectroscopic characterisation as well as the use of modern flow reactor technology. For an example of a past summer project that was subsequently published, please see: https://doi.org/10.1016/j.tetlet.2021.153522
Relevant majors: Chemistry, Chemical Engineering, Medicinal Chemistry, Biology
exploiting flow chemistry for the safe generation and use of novel reactive intermediates
Supervisor: PI: associate professor marcus baumann
A second summer research project is offered that will provide a talented student with the opportunity to acquire and master important research skills in a project at the interface of organic chemistry, flow chemistry, biology and chemical engineering.
This project will also exploit continuous flow chemistry (A field guide to flow chemistry for synthetic organic chemists - Chemical Science (RSC Publishing)) as a novel and exciting addition to the chemist’s toolbox that allows to perform chemistry in a safer, more effective and sustainable manner yielding readily scaled and automated processes that are highly desirable in academia and industry alike.
Specifically, in this project we will apply flow reactors to the synthesis of reactive intermediates (Generation and Use of Reactive Intermediates Exploiting Flow Technology | CHIMIA) that are fleeting and highly unstable molecules with significant potential in organic synthesis. Conventional batch chemistry is not suitable for the scaled generation of reactive intermediates such as carbenes, nitrenes, benzynes and various organometallics due to safety concerns when scaling such reactions to make building blocks on gram scale with relevance to pharma and fine chemical industries. This project will focus on developing new methods using photochemical and cryogenic reactions to generate and consume selected reactive intermediates using miniaturised flow reactors. In addition to synthesising a number of valuable chemical building blocks the student working on this project along with a PhD researcher will purify the final products and use a variety of spectroscopic techniques (NMR, IR, MS) to fully characterise these as needed for subsequent publication of the results.
The successful student will be embedded in our international research group (currently 10 PhDs, 2 postdocs) and gain new skills in chemical synthesis, purification, spectroscopic characterisation as well as the use of modern flow reactor technology. For an example of a past summer project that was subsequently published, please see: https://doi.org/10.1016/j.tetlet.2021.153522
Relevant majors: Chemistry, Chemical Engineering, Medicinal Chemistry, Biology
exploring the future of game development, through the creation of an open-source irish biodiversity game
Supervisor: dr abraham campbell
This an ongoing project investigates how Generative AI can shape the future of game development by helping complete the prototype of the Irish Biodiversity Game. The existing prototype game takes the form of a top-down JRPG with point-and-click mini-games, where players identify animals across Irish biomes such as forests, mountains, and coastal regions.
The student will work with existing project with members to refine the game and help evaluate its use.
This work will be in collaboration with the UCD Biology and Environmental Science School and presents practical coding challenges and an opportunity to explore the future of game development, as well as starting point in exploring the field of serious games where games are designed to not only be fun but also have a real positive impact on the world.
Relevant majors: Computer Science
VR in space; Virtual reality beyond earth: a framework for assessing vr feasibility in space environments
Supervisor: dr abraham campbell
Across three decades of VR and 3D graphics development, we have moved from the pioneering simplicity of Super Mario 64 to the rich fidelity of modern titles like Doom Eternal. On Earth, these leaps were driven by consumer hardware performance, but in space, the equation changes entirely: cosmic radiation threatens the stability of GPUs and CPUs, causing errors, crashes, and even hardware failure.
This ongoing project seeks an intern to develop small 3D examples using UNITY to demonstrate on the framework what 3D worlds are possible and feasible under given condition.
In doing so, the project combines historical perspectives on VR design with forward-looking insights into how virtual environments might support astronaut training, recreation, and psychological well-being beyond Earth.
Relevant majors: Computer Science
Can gut cells sense bacterial invasion? The role of Piezo1 in Streptococcus gallolyticus colonic cell invasion.
Supervisor: Dr. Jennifer Mitchell
The mechanoreceptor Piezo1 senses membrane integrity and when colonic cells are breached by gut pathogens that invade through the cell membrane it triggers ATP release as a DAMP signal to the immune system. Interestingly Streptococcus gallolyticus can invade colonic epithelial cells using twitching motility that is inhibited with excess nucleotides and nucleosides. This project involves testing whether Sgg triggers ATP release in colonic epithelial cells via stimulation of Piezo1 and whether Piezo1 agonists and antagonists can inhibit or promote Sgg invasion of colonic epithelial cells. The student will learn cell and bacterial cell culture, sterile technique, light/fluorescent microscopy and cell invasion assays.
Relevant Majors: Microbiology, Biochemistry, Cell Biology
Investigation of Short-Term Memory using Pre-Questioning Technique
Supervisor: Assistant Professor Fun Man Fung, PhD FICI
This project aims to experimentally determine the effect of a pre-questioning technique (PQ-T) on the efficiency and capacity of human short-term memory (STM), specifically focusing on the encoding and retention of sequential, non-semantic information. We seek to assess whether the act of actively predicting or formulating expectations about an upcoming stimulus prior to its presentation systematically alters the encoding process, leading to measurable differences in subsequent recall performance.
Relevant Majors: Biology, Chemistry, Computer Science, Biomedical Sciences, Environmental Biology, Genetics, Psychology
The Silence of Science: Psychological Safety and Inquiry Reluctance in STEM Education
Supervisor: Assistant Professor Fun Man Fung, PhD FICI
This study aims to investigate the factors contributing to students' reluctance to ask questions in STEM (Science, Technology, Engineering, and Mathematics) learning environments, specifically examining the role of psychological safety (PS). The project seeks to quantify the relationship between perceived psychological safety within the classroom: defined as the belief that one will not be punished or humiliated for speaking up with ideas, questions, concerns, or mistakes; and the self-reported frequency and nature of student inquiry. The ultimate goal is to identify specific pedagogical or environmental variables that foster or inhibit a climate of inquiry.
Relevant Majors: Biology, Chemistry, Computer Science, Biomedical Sciences, Environmental Biology, Genetics, Psychology
Creating a Cybersecurity Dataset for Open RAN Networks
Supervisor: Associate Professor Madhusanka Liyanage
This project introduces the student to the cutting-edge world of mobile networks by creating a unique cybersecurity dataset for 6G Open Radio Access Networks (Open RAN). The student will simulate realistic attack scenarios—such as spoofed signals or manipulated applications—and capture traffic data using a controlled testbed. They will then label and organize this data to support machine learning research in intrusion detection. The final outcome will be a high-quality, publicly shareable dataset that fills a major gap in current mobile network security research. The student will gain hands-on experience in ethical hacking, data analysis, and cybersecurity and work on operation 6G testbed.
Relevant Majors: Computer Science, Data Science, Cybersecurity, Electrical Engineering
Privacy-Preserving AI on Edge Devices with FPGA Acceleration
Supervisor: Associate Professor Madhusanka Liyanage
Large Language Models (LLMs) like ChatGPT require huge resources—but what if they could run on small devices such as IoT sensors or mobile robots? This project allows students to design and test methods for making AI models more efficient and privacy-preserving. By combining Federated Learning and FPGA hardware accelerators, students will explore how to distribute AI training without sharing raw data, keeping user information private. The project introduces key skills in AI, hardware, and privacy protection, while letting students test their solutions on UCD’s advanced 5G/6G research testbeds.
Relevant Majors: Computer Science, Data Science, Electrical Engineering
Immersive Digital Twin of a Mobile Network Testbed
Supervisor: Associate Professor Madhusanka Liyanage
Imagine being able to “step inside” a live mobile network and see how data flows, devices connect, or even how cyberattacks unfold. This project will let students build a digital twin—a virtual copy—of UCD’s 6G Open RAN testbed. By linking real-time telemetry from the network to a 3D environment, students will create interactive visualizations of network operations. The system can simulate failures, visualize security events, and provide training tools for researchers. This project is perfect for students interested in virtual reality, networking, and hands-on software development.
Relevant Majors: Computer Science, Data Science, Computer Engineering, Interactive Media
AI-Powered Privacy Compliance Recommender
Supervisor: Associate Professor Madhusanka Liyanage
With new regulations such as the EU AI Act, ensuring that AI systems are safe and privacy-preserving is critical. In this project, students will design a recommender system powered by Large Language Models (LLMs) to help developers check whether their machine learning models meet privacy requirements. The system will analyse model behaviours, suggest improvements (such as adding differential privacy), and test against real-world use cases. Students will acquire valuable skills in AI regulation, adversarial testing, and ethical AI development—an area of increasing importance for both industry and research.
Relevant Majors: Law, Social Sciences, Computer Science, Data Science, Law & Technology
Real-Time Digital Twin for Smart Vehicles over 6G ORAN
Supervisor: Associate Professor Madhusanka Liyanage
This project provides students with the opportunity to design and build a VR-based digital twin of a smart car. Using a small sensor-equipped RC car as the physical model, students will replicate the car’s movements and sensor data in a VR environment. The system will connect through a low-latency 6G ORAN network, allowing real-time synchronization between the real and virtual vehicles. Applications include autonomous driving, intelligent traffic systems, and IoT analytics. Students will get hands-on experience in VR development, sensor integration, and connected vehicle technology.
Relevant Majors: Computer Science, Electrical Engineering, Data Science, Interactive Media
Reinventing Olefination Reaction using Sustainable Phosphorus Sources
Supervisor: Dr Kirill Nikitin
This project offers an opportunity to join a team pioneering a sustainable transformation of the classic Wittig olefination reaction. The goal is to create a cleaner, waste-free process that avoids hazardous reagents and supports circular phosphorus use.
- Green Chemistry Innovation: The new method reuses the phosphine oxide by-product as a starting material, removing the need for phosphines and reducing chemical waste.
- Eigenbase Reagents: Development of novel ion-pair carboxylate reagents that contain their own built-in base, allowing reactions to proceed without added bases or protecting groups.
- Sustainable Synthesis: The process (1) avoids halogen derivatives, metal salts, and bases, making it environmentally friendly and efficient.
- Catalytic Cycle (2): Exploration of new acid-catalysed circular olefination reactions (CORE) that achieve the transformation in a single step.
- Double catalysis: combine acid catalysis with continuous sustainable recovery of phosphine species (3).
This work supports the SINFERT Project, focused on phosphorus recovery and recycling from industrial and agricultural waste. The research explores how phosphorus-containing by-products from organic synthesis can be reintegrated into industrial phosphorus cycles, linking laboratory innovation with large-scale sustainable resource management.
What You’ll Learn
- Air-free wet chemistry (handling reactions without oxygen or water)
- Advanced NMR spectroscopy (¹H, ¹³C, ³¹P)
- Isolation and purification of organic compounds
- Introductory DFT computational chemistry
- Industrial phosphorus recovery and sustainable process design
Ideal candidates are undergraduates in Chemistry, Medicinal Chemistry, or Chemical Engineering with strong interests in synthesis, catalysis, and green chemistry, and with significant hands-on laboratory experience in experimental chemistry.
Relevant Majors: Chemistry, Medicinal Chemistry, Chemical Engineering
Are nanoparticles better DNA microcarriers for genetic modification of plants cells by biolistic transformation?
Supervisor: Assoc. Prof. Carl Ng
Genetic engineering of plant cells using biolistic transformation has relied on the use of DNA microcarriers made from tungsten or gold. This project aims to evaluate the suitability of nanoparticles as DNA microcarriers for the delivery of transgenes into plant cells, and compare the transformation efficiency of nanoparticles vs gold particles.
Relevant Majors: Plant Biology, Cell & Molecular Biology, Genetics
Genetic basis of vertebrae numbers in an endangered North American cichlid fish
Supervisor: Dr Darrin Hulsey
Vertebrae number is often diagnostic for a given species and cichlid fishes can often vary extensively in this emblematic vertebrate trait. The project will make use of previously collected specimens and x-rays or micro-computed tomography scans to examine population differences in fish vertebrae. Ultimately, this study will combine quantitative trait loci (QTL) mapping, genome wide association using whole genome data, and high throughput phenotyping using micro-computed tomography. These methods will finally be combined to examine the genomic basis of vertebrae numbers in a cichlid fish endemic to the Chihuahuan desert of North America.
Mimicking Nature's Processes: Harnessing Radicals for Catalytic Reactions
Supervisor: Dr Tom Hooper
The catalysis industry produces over $500 billion worth of products worldwide each year and homogeneous catalysts are used to produce a vast range of compounds, from feedstock chemicals to complex drug molecules. Many of these catalysts are based on rare and expensive transition metals and require energy intensive processes. In contrast, nature harnesses the reactivity of unpaired electron species (radicals) to form key biomolecules under mild conditions, using more abundant and less toxic elements. Developing catalysts which can mimic the radicals used in natural processes will make catalytic synthesis less expensive and more sustainable.
The aim of this project is to synthesise and characterise a range of phosphorus compounds with varying properties that readily form radical compounds in solution. This will be done by first forming a scaffold for the phosphorus atom using organic synthetic methods, changing the steric and electronic properties, followed by introduction of the phosphorus centre and reduction to form the target radical compound using standard air sensitive methods. These potential catalysts will be evaluated for reactivity towards small molecules (H2, NH3, CO2 etc.) and organic substrates (alkenes, alkynes, furans, carbonyls etc.). Onward reactivity towards the functionalisation of these substrates will be targeted with the aim of regeneration of the catalytic centre. In situ analysis of these reactions will be performed by multinuclear NMR spectroscopy, electrochemistry and reaction kinetics studies.
Relevant Majors: Chemistry, Biochemistry
Simulations to Elucidate the Impacts of Composition and Nanostructure on Base-Catalyzed Polystyrene Decomposition
Supervisor: Dr Evan Walter Clark Spotte-Smith
Polystyrene (PS) makes up roughly 10% of all plastic waste, making it an attractive target for circular recycling efforts. Although PS can be converted to its monomer (styrene) through thermal decomposition alone, this process is extremely energy-intensive, making it impractical based on both economic and environmental concerns. However, there is another route available: PS can be chemically recycled at modest temperatures using solid base catalysts (e.g., MgO).
In this project, a student will apply molecular dynamics (MD) using "universal" machine learning interatomic potentials to simulate base-catalyzed PS decomposition. The aim of the project is to understand the elementary mechanism(s) underlying the base-catalyzed reaction and to understand how various factors — including surface basicity, defects, and nanostructure — impact the mechanism and associated kinetics. In writing MD workflows and analyzing reactive trajectories, the student working on this project will gain experience in thermal catalysis, chemical kinetics, and computational chemistry. Collaboration with experimental partners is also possible.
Students from all backgrounds are welcome to engage in this project. Prior experience with computer programming (especially using Python or Julia), high-performance computing, and command-line interfaces (especially using Linux operating systems) would be beneficial but are not required.
Relevant Majors:Chemistry, Chemical Engineering, Materials Science, Physics
Analysis of Exogenous RNA From Probiotic Bacteria
Supervisor: Dr Paola Valentini
Probiotic bacteria influence our well-being in several ways. An increasingly recognised one is through the delivery of extracellular vesicles (EVs), tiny particles surrounded by a membrane and containing a diverse cargo of nucleic acids, proteins and peptides. Among these molecules are several species of RNA. Such RNA molecules likely have a role in how bacteria interact with the environment and with the host, as they are specifically encapsulated in the vesicles, the content of the latter being different from that of the intracellular milieu. However, the functions of most of these RNAs remain elusive. The project aims to study the presence of particular RNA species in EVs RNA by reverse-transcription PCR (RT-PCR) and quantitative reverse-transcription PCR (qRT-PCR). This project can be accomplished by three students, each working independently on different variables, different RNA targets, or different bacterial strains.
Skills and techniques training list:
- General microbiology techniques (bacterial cell culture, preparation of media and glycerol stocks)
- Culturing anaerobic bacteria
- Isolation of extracellular vesicles (EVs)
- Working with RNA
- PCR, RT-PCR, qRT-PCR.
The student will also have the chance to confront particular technical issues related to EVs purification and RNA extraction from non-model bacteria.
Relevant Majors: Cell Biology, Microbiology
Association of Lipopolysaccharide Binding Protein (LBP) Gene Expression with Colorectal Cancer
Supervisor: Dr. David Hughes
Colorectal cancer is a common cancer with currently alarming increases at younger ages, making prevention highly important. Although there are complex disease causes, environmental factors, particularly obesity and lifestyle, are known to play a strong role. Recent evidence suggests that commensal microbial dysregulation and exposures to microbial toxins are involved in its development. We hypothesis that this occurs through inflammatory-induced weakening of the protective gut mucosal barrier by obesity, dietary/lifestyle, and microbiome factors that lead to translation of pathogenic bacteria and their toxins into the epithelial lining of the gastrointestinal tract. To help explore this hypothesis, the student will measure a biomarker of gut-barrier function and bacterial translocation (indicating microbial dysbiosis). This will be done by measuring blood and tumour tissue derived RNA expression analysis of the human Lipopolysaccharide Binding Protein (LBP) for bacterially derived lipopolysaccharide – integral to an intact barrier (which modulates the permeability of tight junctions between intestinal tract cells). These will be measured in a series of blood samples from non-disease controls (n=40) and patients with colorectal cancer (n=40), while a subset of the latter used to measure the tumour and matched normal tissue expression of LBP).
Secondly, existing data on common genetic variants (SNPs; single nucleotide polymorphisms) in human genes within the LPS / LBP pathway (see our related study at doi: 10.1093/mutage/geae008) will be correlated with the observed gene expression from these same patients. Together, these findings will illuminate gut-barrier function in colorectal cancer and possible contribution of bacterial toxin exposure from the gut. Note that both Postdoctoral and PhD researchers in the lab will be available to provide support to students who are unfamiliar with these methods. By the end of the project the student will become familiar with techniques including RNA extraction and gene expression assays, and biostatistical packages such as R.
Relevant Majors: Microbiology, Genetics, Biochemistry, Epidemiology, Biology, Biostatistics
Targeting Oxidative Stress and Prolyl Hydroxylase Domain Inhibition as Neuroprotective Strategies in an Oxygen Glucose Deprivation (OGD) Model in Rat Hippocampal Slices.
Supervisor: Professor John O’Connor, SBBS
During hypoxia several physiological changes occur within neurons including the stabilization of hypoxia-inducible factors (HIFs). The activity of these proteins is regulated by O2, Fe2+, 2-OG and ascorbate dependant hydroxylases which contain prolyl-4-hydroxylase domains (PHDs). PHD inhibitors have been widely used and have been shown to have a preconditioning and protective effect against a later and more severe hypoxic insult. In addition, oxidative stress plays an important role in ischemia-reperfusion injury (IRI). Antioxidants have been shown to have beneficial effects during increased levels of reactive oxygen species in ischemic stroke. Therefore, this study will investigate the effects of antioxidants, SOD mimetics and novel PHD inhibitors on synaptic transmission and neuronal viability in an oxygen-glucose deprivation (OGD) rat stroke model. The project will involve learning the technique of electrophysiology and recording electrical activity from isolated rat brain slices. Excitatory post-synaptic potentials (fEPSPs) will be elicited by stimulation of the Schaffer collateral pathway in young rat CA1 hippocampal slices. During OGD hippocampal slices will be perfused with glucose-free aCSF bubbled with 95% N 2 /5% CO 2 for 20 min. Synaptic plasticity will be measured as long-term potentiation following the oxygen glucose deprivation sand assessed. Ultimately the project seeks to identify agents that can protect neurons and synapticity plasticity during and post stroke-like symptoms.
Relevant Majors: Neuropharmacology, Physiology, Neuroscience
The Evolution of Climate Adaptation Genes in Frogs
Supervisor: Associate Professor Katharina Wollenberg Valero
Amphibians are the most threatened group of vertebrates, and we urgently need to understand better how they respond to climate change stressors such as high temperatures. Our previous research has shown that adaptation to temperature in vertebrates is controlled by a limited set of genes, but we don’t yet know much about this phenomenon in frogs. In this project, you will conduct a bioinformatic study comparing sequences of these genes between species of frogs, and correlating these genetic differences with the climatic conditions they live in, to further narrow down which genes we should study when assessing the risk of frog species to changing climates.
Skills and techniques training: Bioinformatics, Alignments, selection analysis.
Relevant Majors: Biology & Environmental Science
The Effect of Temperature on Multicellular Development in Zebrafish Embryos
Supervisor: Associate Professor Katharina Wollenberg Valero
This project will investigate how early development is affected in zebrafish embryos undergoing thermal stress. You will study embryos at the timepoint of gastrulation, where ectoderm, mesoderm and endoderm are forming and determine the expression of candidate genes involved in the formation of these cell layers. This will shed light on the effect of heat stress on cell-to-cell communication in multicellular organisms. If there is time and scope we can also expand the project towards chimera assays which would involve the mixing of fluorescently labeled cells from heat stressed vs control embryos to study cellular competition.
Skills and techniques training: working with embryos, microscopy, LAMP gene expression, possibly microinjection.
Relevant Majors: Biology & Environmental Science
Demonstration of a Stereocontrolled Method to Prepare 1,3-Dienes
Supervisor: Drs Karthik Manoharan and Paul Evans
Conjugated 1,3-dienes are useful structural motifs (e.g. 1 in Figure 1). They are present in several
naturally occurring compounds such as the pheromone bombykol and participate in important
reactions such as the Diels-Alder cycloaddition. We aim to prepare this valuable unit in a new manner.
Specifically, we will combine the Horner-Wadsworth-Emmons (HWE) reac-on of sulfonyl phosphonate
2 to prepare vinyl sulfone 3. Previous work in our group2 has demonstrated that compounds of the
type 3 (known as vinyl sulfones) undergo alkene isomerisation to generate allylic sulfones 4 and that,
depending on the conditions employed, either the Z- or E-alkene isomer can be accessed. We then
plan to intercept the anion from 4 in a Julia-Kocienski (JK) olefination reaction to access 1. Notably,
since all reactions proceed under basic condi-ons, we are confident that this ambitious sequence can
be executed in one reaction flask, thereby improving the efficiency of the process.3
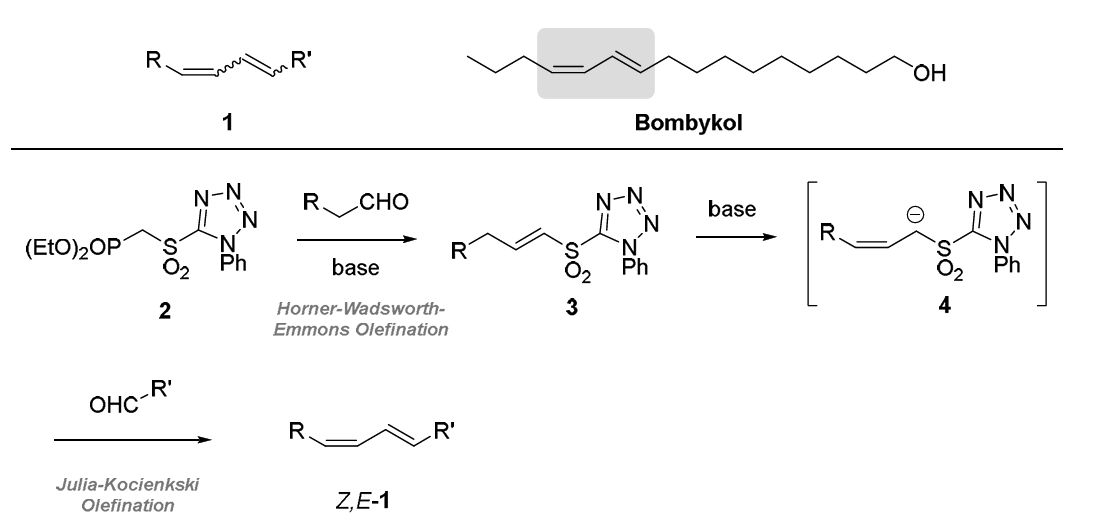
Figure 1. Conjugated 1,3-dienes and their proposed synthesis via a new HWE-JK cascade sequence.
This project will be housed in the Evans chemistry laboratory in the School of Chemistry in UCD and
the work will be underpinned by the UCD NMR spectroscopy facility. This lab currently has three
postgraduate researchers working on projects related to this proposal. People with an interest in
laboratory work, synthetic organic chemistry and who have some general undergraduate chemistry
(Gen Chem and OChem) laboratory experience would be particularly suited to the proposed project.
References
1. P. Mauleon, A. Alonso, M. Rodríguez Rovero and J. C. Carretero, J. Org. Chem. 2007, 72, 9924-
9935.
2. Unpublished work. For a report related to this proposal see: N. Collins, R. Connon, G. Sánchez-
Sanz and P. Evans, Eur. J. Org. Chem. 2020, 6228-6235.
Large Language Models (e.g., ChatGPT/BERT/LLaMA) and Agentic Artificial Intelligence for Digital Forensic Investigation
Supervisor: Assoc. Prof. Mark Scanlon
A large language model (LLM) is a type of artificial neural network designed for understanding and generate human-like text. It is a subset of models within the broader domain of natural language processing (NLP), which focuses on enabling machines to interpret, generate, and respond to human language. This project aims to develop a novel approach for digital forensic investigation using language models, such as ChatGPT/BERT/LLaMa, to generate forensically sound solutions for discovering and analysing digital evidence. The proposed method will evaluate the potential for LLMs to enable digital investigators to analyse and interpret digital evidence from a variety of digital data sources and devices. Specifically the project will look at the prospect of an agentic AI approach to enable a natural language investigative query being taken as input, a step by step process being generated to answer the query with a variety of agents, automating the agentic evidence discovery through automatically generated scripting, interpreting the results and presenting this interpretation back using natural language. The solution will be benchmarked using AutoDFBench a benchmarking framework designed in my group.
The work of the summer research student would involve working in a team of researchers in the fine-tuning and testing of AI agents contribute towards artificial intelligence aided digital forensic investigation. Working with this ongoing project in my research group will expose the research student to the full research lifecycle from research design, development, experimentation with devices and datasets, and dissemination/discussion of results at UCD Forensics and Security Research Group meetings.
Relevant Majors: Computer Science, Computer Engineering, Data Science, Software Engineering
Computer Vision Based Indoor Multimedia Geolocation
Supervisor: Associate Professor Mark Scanlon
The task of multimedia geolocation is becoming an increasingly essential component to effectively combat human trafficking and other criminal activity. While text-based metadata can easily provide geolocation information with access to the original media, this metadata is stripped when shared via social media and common chat applications. Geolocating, geotagging, or finding geographical clues in the multimedia content itself is a complex tax. While there are numerous manual/crowdsourcing approaches to this, recent research has shown that computer vision is one viable avenue for research.
The work of the summer research student would involve working in a team in the creation of novel datasets for multimedia geolocation and developing computer vision-based techniques for indoor multimedia geolocation. The aim is to develop powerful methods for image geolocation that enable more efficient investigations in the field of human trafficking. Colour values serve here as a key component to describe specific characteristics of an image and colour-based descriptors will be used for Content-Based Image Retrieval. The performance of the developed methods will be evaluated using the Hotels-50K dataset as a foundation. Working with this ongoing project in my research group will expose the research student to the full research lifecycle from research design, development, experimentation with devices and datasets, international collaboration, and dissemination/discussion of results at UCD Forensics and Security Research Group meetings.
Relevant Majors: Computer Science, Computer Engineering, Data Science, Software Engineering
Leveraging Large Language Models' Semantic Space Mapping to Overcome Encryption for Law Enforcement
Supervisor: Assoc. Prof. Mark Scanlon
Passwords have been and still remain the most common method of authentication in computer systems. From accessing your smartphone, to setting up your online banking account or social identification, there is a plethora of passwords that users are required to set and remember in hundreds of websites. Complex passwords make the job of law enforcement engaged in a digital investigation more difficult, especially since time is often of the essence.
This project aims to provide insights into the password selection process of users and the impact of contextual information in it. Additionally, the ways that this contextual information can be leveraged in order to assist with the lawful password cracking process will be explored. Publicly released Large Language Models (LLMs) are trained on as much data as possible and semantically map words. This project aims to exploit this semantic mapping to generate better context based password candidate dictionaries. To this end, intelligent, community-targeted dictionaries will be assembled on a variety of topics. For example, when targeting a community of superhero enthusiasts, we assume and want to prove that a larger proportion of passwords would be contextually relevant to that community than not.
The work of the summer research student would involve contributing to the generation and evaluation of existing and newly created bespoke “smart” context-based dictionaries against existing leaked password datasets. The tools and dictionaries will be evaluated for different metrics and for varying degrees of contextualisation, with the aim of establishing the impact of context in passwords. Additionally, the summer research student will be able to contribute ideas to the dictionary creating process and be able to test their own dictionaries vs. existing dictionary-based approaches with this framework.
Relevant Majors: Computer Science, Computer Engineering, Data Science, Cybersecurity
Using Zebrafish to Understand if Mutations in a Visual Cycle Gene are Disease Causing.
Supervisor: Professor Breandán Kennedy
The aim of this project is to understand the fundamental biology of the visual cycle and to evaluate the pathogenicity of RGR patient variants associated with vision loss using zebrafish. For vision to be maintained, there needs to be constant recycling of the visual chromophore 11-cis-retinal (11cRAL), which is a light-sensitive form of vitamin A. The light-induced production of 11cRAL is mediated by the visual cycle gene RGR (reviewed by (1)). Mutations in RGR are linked to retinal diseases such as such as inherited retinal degeneration (IRDs) (2), a group of eye disorders which are a leading cause of blindness and age-related macular degeneration (AMD) (3). There are limited treatment options available for individuals with IRDs and AMD.
There are very few confirmed RGR variants that are disease causing. A small tropical fish known as the zebrafish is being used to determine if specific RGR patient variants are disease causing. The zebrafish eye structure is similar to a human eye. Behavioural assays such as the optokinetic response (OKR) (4) or visual motor response will be used to determine if visual functional is impacted because of mutations in RGR.
In this project, the student will be trained in zebrafish husbandry. They will potentially learn techniques associated with microinjection and the screening of zebrafish larvae. Using behavioural tests such as the OKR, the student will also be involved in characterising the functional vision of zebrafish expressing RGR variants. The retinoid profiling (quantification of 11cRAL) of zebrafish larvae may be potentially investigated also.
- Ruddin G, McCann T, Fehilly JD, Kearney J, Kennedy BN. The dark and bright sides of retinal G protein-coupled receptor (RGR) in vision and disease. Prog Retin Eye Res. 2025 Feb 15;106(101339):101339.
- Morimura H, Saindelle-Ribeaudeau F, Berson EL, Dryja TP. Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat Genet. 1999 Dec;23(4):393–4.
- Jiang M, Shen D, Tao L, Pandey S, Heller K, Fong HK. Alternative splicing in human retinal mRNA transcripts of an opsin-related protein. Exp Eye Res. 1995 Apr;60(4):401–6.
- Gómez Sánchez A, Álvarez Y, Colligris B, Kennedy BN. Affordable and effective optokinetic response methods to assess visual acuity and contrast sensitivity in larval to juvenile zebrafish. Open Res Eur. 2021 Aug 12;1:92.
Relevant Majors: Neuroscience, Genetics, Pharmacology
Improving the Stability and Bioactivity of the Therapeutic Protein Human Interferon-Gamma
Supervisor: Dr. Marina Rubini
The spread of drug-resistant pathogens is one of the most concerning threats for public health, which is even more acute due to the absence of a clinical pipeline of new antimicrobials. In the face of this global challenge, it is critical to develop new therapeutic approaches. A notable candidate is human Interferon-gamma (hIF-gamma), a glycosylated therapeutic cytokine that belongs to the interferons, a class of proteins that play a crucial role in host defense response to viral and bacterial infections.
In our lab, we selectively modify recombinantly produced (in Escherichia coli) Interferon-gamma variants containing cysteines (Cys) at the naturally occurring glycosylation sites, by exploiting the reactivity and versatility of thiol groups of Cys. After protein purification by affinity chromatography, different defined synthetic glycans or polyethyleneglycol (PEG) moieties are conjugated to the protein variants through a range of thiol-selective bioconjugation with the aim of a) understanding the role of glycans on biological activity and b) improving the half-life of the protein for therapeutic applications. After protein purification by size-exclusion chromatography, we characterize our modified proteins (mass spectrometry, circular dichroism, UV-VIS spectroscopy) and we will assess their stability against proteolysis and unspecific aggregation as well as their bioactivity.
Relevant Majors: Chemistry, Biochemistry
Agent-Based Stock Market Modeling
Supervisor: Dr. Vivek Nallur
The field of Finance has seen enthusiastic adoption of AI technologies. These have mostly been used to model asset pricing as well as trading behaviour, under various conditions. The behaviours seen, as well as the aggregate effects on the markets, have been under assumptions of rational or adaptive traders. The arrival of LLMs have brought with them the possibility of mimicking real human behaviour. However, the size of LLMs make it prohibitive to use them as individual agent-decision drivers.
Small Language Models (SLMs) like Llama 3.1 8B, Qwen2, Phi-3.5, Mistral Nemo, etc. provide the potential to overcome this handicap. The student will create agent-based models, that use SLMs in their agent descriptions or decision-making, to investigate market dynamics, such as bubbles, crashes, and volatility. This will involve creating python-based agents, which interact with SLMs, to drive their behaviour.
Relevant Majors: Computer Science, Computational Social Science, Data Science, Behavioral Economics
Agent-Based Pandemic Modeling
Supervisor: Dr. Vivek Nallur
The field of health and epidemiology is concerned with modelling, both the spread of disease as well as interventions and their impact. Large-scale modelling is essential for policy makers to understand the possible impact of various policies on individual behaviour (which itself can be modelled from simple rules to complex adaptive decisions).
Small Language Models (SLM) like Llama3.1 8B, Qwen2, Phi-3.5, etc. allow modellers to use the nuance present in LLMs, but still be able to run them on individual laptops. This project will experiment with modelling disease spread, and validate if Small Language Models can produce realistic behaviour, both at the individual level as well as the aggregate level. The student will implement agent-based models, which comprise SLMs, using python-based toolkits.
Relevant Majors: Computer Science, Computational Social Science, Data Science, Behavioral Economics
Agent-Based Modeling of Cultural Norms
Supervisor: Dr. Vivek Nallur
Norm emergence occurs in almost all socio-technical systems. However, the mechanisms underlying this emergence are not very well-understood. The presence or absence of top-down rules or constraints on behaviour at the bottom-up level can both contribute to the emergence and sustenance of norms. A systematic experimentation with realistic behaviour from human-like agents would provide some insights into this process. However, LLMs are extremely expensive, in terms of resources, to be deployed for multiple agents.
The use of Small Language Models (SLMs) like Llama 3.1 8B, Qwen 2, Phi-3.5, Mistral Nemo, etc. which can be run on laptops, has been proposed as a possible solution. The student will implement agent-based models that leverage SLMs for agent description as well as decision-making, to investigate the phenomena of norm emergence.
Relevant Majors: Computer Science, Computational Social Science, Data Science
NADH-Inspired Photodynamic Agents for Targeted Release of Radicals
Supervisor: Dr Bartosz Bieszczad
The role of radicals in biological systems is a subject of intensive research. Far from being merely detrimental by-products, radical species are key intermediates in biosynthetic pathways, energy metabolism, redox processes, cell signalling, and defence against pathogens. However, their medical use is largely restricted to a "healing-by-killing" approach, such as in photodynamic therapy (PDT), where reactive oxygen species damage cancerous tissue.
Constructive medical applications of radicals remain rare and underexplored. The most prominent exception is nitric oxide, used to treat angina. Less electronegative, carbon-centred radicals, such as endogenous acyl and ketyl radicals, also show promise for non-destructive applications, including potential treatments for Alzheimer's disease and tuberculosis. Using radicals to finely tune biochemical processes, such as intercepting cell communication, could significantly impact therapeutic strategies. However, their reactive nature makes controlled generation challenging. A key obstacle is the scarcity of chemical agents capable of selectively generating desired radical species within a biological environment.
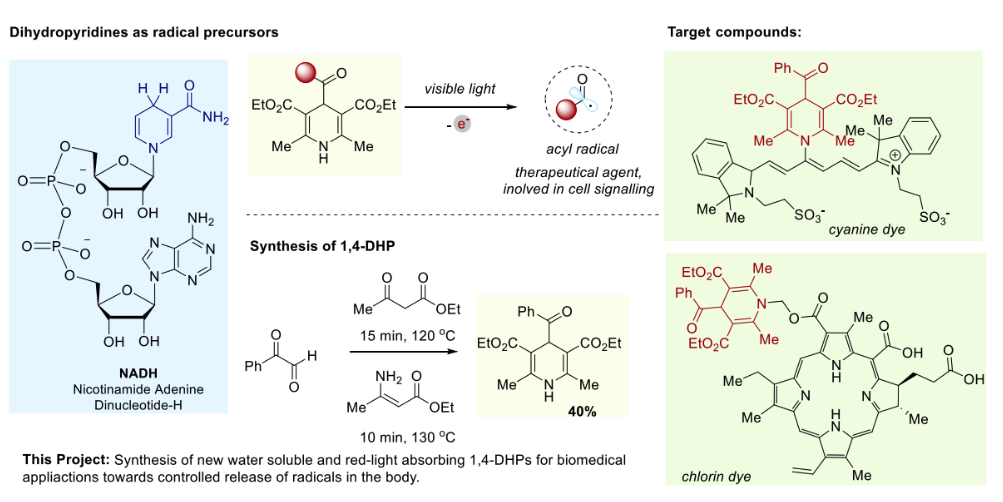
Scheme 1. Project outline: (a) 1,4-DHP as radical precursors; (b) Synthesis of DHPs; (c) Proposed new
photodynamic agents, utilizing water soluble and red-light absorbing chromophores.
Inspired by nature, we propose using NADH analogues—1,4-dihydropyridines (1,4-DHPs)—as radical precursors for PDT. These molecules can be easily synthesized in a single step and release alkyl and acyl radicals upon irradiation. To adapt them for PDT, they must be water-soluble and absorb red light. This project aims to prepare hybrid 1,4-DHP molecules that meet these crucial criteria. The student will be involved in the design and synthesis of new 1,4-DHP hybrids. The products will be characterized using NMR and UV-Vis spectroscopy, and their radical-releasing properties will be tested using a reaction with a radical trap.
Relevant Majors: Chemistry, Biochemistry
Using Camera Trap and Acoustic Surveys to Reveal the Dynamics of Deer “Hotspots” in Ireland
Supervisor: Dr Simone Cuiti and Dr Colin Brock
Camera trap and passive acoustic monitoring surveys are important for gathering information on terrestrial wildlife’s distribution and behaviour, providing insights into ecosystem health and functioning. These methods play a crucial role in informing conservation strategies and wildlife management decisions. The UCD Laboratory of Wildlife Ecology and Behaviour has established two major Irish projects monitoring wildlife using non-invasive techniques. The first one - Snapshot Europe - is a coordinated camera trap effort to collect data on mammals across Europe. The second one – bioDEERversity – is a govt-funded biodiversity monitoring program set up in the Wicklow and Dublin mountains.
These projects have been gathering data on wildlife and general biodiversity (soil ecology, plant diversity, and vertebrate diversity) in a deer hotspot area of Ireland, where Sika deer have been shown to occur at unsustainable high densities. Camera traps and acoustic recorders have been set up to capture data in areas with no deer (fenced exclusion zones) as well as in those areas spread across a gradient of sika deer relative density. Students involved will learn how to use user-friendly deep learning softwares (e.g., DeepFaune and Kaleidoscope Pro) designed to identify species using large numbers of image and audio files. Furthermore, the students will analyse camera trap data and/or acoustic data, and will have the opportunity to tackle a research question that will be defined with the help of the supervisor (e.g.: What is the effect of deer presence on the occurrence and diversity of the other mammal species?).
Relevant Majors: Biology, Environmental Science
Glycoconjugate for Targeting Bacterial Carbohydrate-Binding Proteins
Supervisor: Dr. Joseph Byrne
Many pathogens, such as bacteria P. aeruginosa and E. coli or fungus C. albicans, produce characteristic carbohydrate-binding proteins. These pathogens are designated as ‘Critical Priority’ by the WHO, requiring new targeting strategies. We have recently shown that luminescent glycoconjugate–lanthanide complexes can detect this class of proteins by characteristic changes in luminescence. You synthesize novel glycoconjugate molecules with suitable sugar motifs to interact with these proteins, learning skills in carbohydrate synthesis, purification, spectroscopy and possibly coordination/supramolecular chemistry. These new compounds will expand a library of glycoconjugates and some will be tested for protein sensing and/or antimicrobial behavior of these materials either in the School of Chemistry, or with collaborators. Some experience with organic synthesis and purification required.
Further reading: K Wojtczak, E Zahorska, IJ Murphy, F Koppel, G Cooke, A Titz, JP Byrne*, “Switch-on luminescent sensing of unlabelled bacterial lectin by terbium(III) glycoconjugate systems”, Chem. Commun., 2023, 59, 8384; doi:10.1039/D3CC02300A ;
K Wojtczak et al. “Antiadhesive glycoconjugate metal complexes targeting pathogens Pseudomonas aeruginosa and Candida albicans”. Org. Biomol. Chem. 2025, Advance Article; doi:10.1039/D5OB00970G)
Assessing the Impact of Room Impulse Responses on the Quality of Live Music Simulations
Supervisor: Dr Andrew Hines, Dr Carl Timothy Tolentino
Music quality is influenced by the listening environment. Room impulse responses allow us to virtually reproduce acoustic signals as if you are physically located inside a room, e.g. the amount of echo and reverberation is different in a small booth to a large theatre. Measuring quality in different environments is costly and time-consuming. We want to assess whether different room impulse responses have an effect on the perceived audio quality of virtual live music signals. The student will be tasked to simulate live music signals with an existing data set, e.g. MUSDB [1], using an existing software, Binamix [2], with different room impulse responses from an existing data set [3]. They will assess the perceived quality of these signals by conducting subjective MUSHRA listening tests with ~10 people using the Go Listen audio evaluation platform [4].
[1] MUSDB18 - a corpus for music separation https://zenodo.org/records/1117372
[2] D. Barry, D. S. Panah, A. Ragano, J. Skoglund, and A. Hines, “Binamix - a python library for generating binaural audio datasets,” in AES Europe Convention 158, vol. AES Convention: 158, Audio Engineering Society, May 2025. https://aes2.org/publications/elibrary-page/?id=22870
[3] https://github.com/ShanonPearce/ASH-Listening-Set/tree/main/BRIRs
[4] https://golisten.ucd.ie
Relevant Majors: Computer Science, Electrical Engineering, Mathematics
Antibiotic Resistance and Phenotypic Changes in the Human Pathogen Campylobacter Jejuni
Supervisor: Assoc Prof Tadhg Ó Cróinín
Campylobacter jejuni is the most prevalent cause of foodborne bacterial gastroenteritis world-wide and the world health organisation has classified it as a pathogen of concern due to the increasing numbers of strains with resistance to fluoroquinolone antibiotics. We recently published a paper which highlighted that fluoroquinolone resistant strains of C. jejuni appear to have different phenotypes to those of the parent strain including increased virulence and decreased motility. This project will focus on the comparison of antibiotic resistant and sensitive strains using a variety of assays including motility assays, invasion assays as well as molecular characterisation of the genetic mutations which cause this resistance. As part of a collaboration with UCDavis strains isolated from a variety of different sources will be used to select for fluoroquinolone resistant isolates which will be assessed using these assays. Methods used will be similar to those outlined in our recent paper on acquisition of fluoroquinolone resistance and its relationship with biofilm formation and pathogenicity. Students interested in taking this project should be highly motivated and interested in the field of infection biology, antimicrobial resistance and understanding how bacterial pathogens cause disease.
Whelan MVX, Ardill L, Koide K, Nakajima C, Suzuki Y, Simpson JC, Ó Cróinín T. Acquisition of fluoroquinolone resistance leads to increased biofilm formation and pathogenicity in Campylobacter jejuni. Sci Rep. 2019 Dec 3;9(1):18216.
Relevant Majors: Biology, Microbiology, Biochemistry
ZOVD-Studio: Zero-Shot Open-Vocabulary Object Detection
Supervisor: Dr Soumyabrata Dev
This mini engineering project delivers a toolkit and mini-benchmark for zero-shot, open-vocabulary object detection while intentionally keeping the approach method-agnostic. The intern may either design an approach using supervised or unsupervised training, or build a training-free detection pipeline with optional few-shot calibration using any combination of modern foundation models (e.g., DINO, CLIP, BLIP, OWL or similar). We will provide: (i) a small evaluation subset; (ii) standard metrics - query mAP@0.5 for images and phrase-grounding top-k hit rate; (iii) latency/FLOPs logging. Expected capabilities: Given an image or a short video and a user-provided list of category names or phrases, the system returns labeled bounding boxes with confidence scores. The demo supports prompt editing, image/video loading, parameter settings, and visualization. No large-scale training is required; few-shot calibration is optional.
Deliverables: a reproducible scripts, unit tests for I/O and evaluation, and a 2–3-page technical note with a small leaderboard (quality vs. latency). The primary goal is a reusable baseline that lowers the barrier to open-vocabulary detection experiments
Relevant Majors: Computer Science
Pruning and Quantization-Aware Training for Efficient Solar Forecasting
Supervisor: Dr Soumyabrata Dev
This project explores how deep learning models can be made smaller and faster without losing accuracy in predicting solar irradiance, an important step toward energy-efficient AI for renewable energy forecasting. The intern will help design and train compact models using pruning and quantization-aware training, which are techniques to reduce model size and computation cost. The work may involve either image-based or time-series-based forecasting pipelines, depending on the student’s interest and background. Applicants should have a basic understanding of Python and deep learning (e.g., using PyTorch or TensorFlow). Prior experience with pruning or quantization is not required; we are looking for curious, motivated students who are eager to learn.
Learning Objectives:
- Learn how solar forecasting models are built and evaluated.
- Understand the basics of model compression and why it matters for efficient AI.
- Get hands-on experience with pruning-aware and quantization-aware training in PyTorch.
- Explore how to balance accuracy and efficiency in real-world deep learning systems.
Relevant Majors: Computer Science, Electrical Engineering, Environmental Engineering
Seeing, Feeling, and Telling: A Unified Framework for Controllable, Trustworthy, and Emotion-Aware Image Captioning with MLLMs
Supervisor: Dr Soumyabrata Dev
This project proposes a unified image captioning framework that combines factual grounding, audience control, and affective sensitivity to improve how multimodal large language models (MLLMs) describe visual content. It integrates two complementary ideas into a single pipeline. First, a See–Plan–Write process enhances reliability and user alignment: the system detects objects and attributes before captioning (See & Check), lets users define goals, tone, and reading level (Plan the Style), and generates accountable text that signals uncertainty and links claims to image regions (Write with Accountability). Second, an Affective Language module interprets low-level visual cues; such as color balance, luminance contrast, stroke dynamics, and texture into emotion profiles grounded in art-psychology priors, which then guide language generation to reflect mood and atmosphere. By conditioning MLLMs on both factual evidence and emotional context, the framework produces captions that are not only accurate and transparent but also expressive and audience-aware. This dual emphasis makes it suitable for diverse applications, from accessible descriptions for visually impaired users to educational tools and art interpretation. The result is a practical, modular system that advances image captioning beyond object recognition toward nuanced, controllable, and trustworthy visual storytelling.
Relevant Majors: Computer Science, Digital Humanities
3D Reconstruction-Driven Digital Twin for Intelligent Network Simulation
Supervisor: Dr Soumyabrata Dev
This project develops a spatially-aware digital twin that integrates real-time 3D reconstruction from SLAM (Simultaneous Localization and Mapping) with intelligent network simulation. Traditional network simulators overlook the impact of real-world geometry such as obstacles, terrain, and spatial layout on communication performance. By reconstructing realistic 3D environments from UAV or ground imagery and linking them to simulated network models, the system enables analysis of location-dependent network dynamics, signal propagation, and adaptive routing in physically grounded conditions. The outcome is a unified framework that connects vision-based spatial understanding with autonomous network optimization, paving the way for more realistic and intelligent digital twin systems.
Preferred Student Profile:
- Background in Computer Science, Electrical/Computer Engineering, or Robotics.
- Skilled in Python or C++, with knowledge of SLAM and basic networking concepts.
- Enthusiasm for AI-driven, interdisciplinary research combining 3D vision and communication systems.
Relevant Majors: Computer Science, Electrical Engineering
Identifying Candidate Space in a Real-Time Scene
Supervisor: Dr Soumyabrata Dev
In the evolving landscape of digital advertising, replacing existing adverts in video frames with dynamic, scene- aware content has become a promising approach. This project focuses on identifying candidate spaces in real-time video scenes where new advertisements can be seamlessly embedded. The primary challenge lies in ensuring the implanted advert blends naturally within the scene, maintaining perspective, scale, and lighting consistency, especially critical in fast-paced environments like sports broadcasts. Several methods have explored advert detection and replacement in image frames[1], but accurate candidate space detection remains a bottleneck for automation and realism. This project aims to develop a machine learning pipeline that detects potential advert zones in video frames by analyzing spatial and contextual cues. The solution must ensure that the selected candidate regions align with the viewpoint and maintain visual plausibility [2]. To train and evaluate the models, we will use established datasets which offer diverse video content for robust analysis. The project will involve data preprocessing, feature extraction, and model development using Python, leveraging state-of-the-art computer vision techniques. This work has strong applications in sports media, digital advertising, and augmented reality, providing a scalable solution for automated advert integration in live video content.
Dataset: CASE [3], SoccerNet [4], Youtube [5]
Project type: Data analysis, Machine Learning, Deep learning Language: Python
References:
[1] Dhang S, Zhang M, Dev S (2025) LaBINet-An approach for seamlessly integrating new advertisements into an existing scene. IEEE Transactions on Artificial Intelligence 6(8):2281 – 2290
[2] Dev S, Javidnia H, Hossari M, et al (2019). Identifying candidate spaces for advert implantation. In: 2019 IEEE 7th International Conference on Computer Science and Network Technology (ICCSNT), IEEE, pp 503–507
[3] S. Dev, M. Hossari, M. Nicholson, K. McCabe, A. Nautiyal, C. Conran, J. Tang, W. Xu, and F. Pitié, The CASE Dataset of Candidate Spaces for Advert Implantation, Proc. International Conference on Machine Vision Applications (MVA), 2019.
[4] Magera F, Hoyoux T, Barnich O, et al (2024) A universal protocol to benchmark cam- era calibration for sports. In: IEEE International Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), CVsports, Seattle, Washington, USA
[5] Abu-El-Haija, S., Kothari, N., Lee, J., Natsev, P., Toderici, G., Varadarajan, B. and Vijayanarasimhan, S., 2016. Youtube-8m: A large-scale video classification benchmark. arXiv preprint arXiv:1609.08675.
Relevant Majors: Computer Science, Electrical Engineering
Probing the Logical Limits of LLMs via Evidence-Grounded Rule Reasoning: A Yu-Gi-Oh! Case
Supervisor: Dr Soumyabrata Dev
Large (vision-)language models often falter on procedural, multi-step rule reasoning, yielding hallucinations or brittle behavior [4, 3, 2]. We propose an evidence-grounded benchmark centered on Yu-Gi-Oh! official materials, where Problem-Solving Card Text (PSCT) defines explicit condition–activation–resolution syntax and official timing guides partition the Fast Effect and Damage Step windows. From these sources, we will curate 300–500 multiple-choice/judgment items with short supporting snippets and links, covering (i) rule-text understanding (PSCT), (ii) multi-step reasoning across target/state changes, and (iii) rule execution to reach correct rulings. A small computer-vision subtask will extract card/state text from official screenshots using open OCR/transformer baselines to enable end-to-end analysis. We will release dataset splits, a dataset card, and minimal evaluation (accuracy, macro-F1, evidence-consistency; optional self-consistency decoding). By anchoring labels in official documents, this project provides a compact, reproducible probe of models’ logical limits on procedural rules, complementing broad benchmarks while minimizing ambiguity and subjective adjudication. We suggest this paper for reading [1].
References
[1] Duan, J., Zhang, R., Diffenderfer, J., Kailkhura, B., Sun, L., Stengel-Eskin, E., Bansal, M., Chen, T., and Xu, K. Gtbench: Uncovering the strategic reasoning limitations of llms via game-theoretic evaluations, 2024.
[2] Li, Y., Du, Y., Zhou, K., Wang, J., Zhao, X., and Wen, J.-R. Evaluating object hallucination in large vision-language models. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing (Singapore, Dec. 2023), H. Bouamor, J. Pino, and K. Bali, Eds., Association for Computational Linguistics, pp. 292–305.
[3] Rawte, V., Chakraborty, S., Pathak, A., Sarkar, A., Tonmoy, S. T. I., Chadha, A., Sheth, A., and Das, A. The troubling emergence of hallucination in large language models - an extensive definition, quantification, and prescriptive remediations. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing (Singapore, Dec. 2023), H. Bouamor, J. Pino, and K. Bali, Eds., Association for Computational Linguistics, pp. 2541–2573.
[4] Sahoo, P., Meharia, P., Ghosh, A., Saha, S., Jain, V., and Chadha, A. A comprehensive survey of hallucination in large language, image, video and audio foundation models, 2024.
Relevant Majors: Computer Science, Artificial Intelligence, Machine Learning, Data Science, Software Engineering
Optimized Crop Type Classification for Adaptive RGB to NIR Image Generation Pipeline
Supervisor: Dr Soumyabrata Dev
Near-infrared (NIR) imagery plays a vital role in plant monitoring and phenotyping, as it reveals physiological and structural details that are invisible in standard color images.
However, acquiring NIR images is often expensive and technically demanding compared to capturing conventional RGB (red, green, blue) images. Consequently, generating NIR images from RGB inputs using deep learning models has become an active area of research, particularly in agricultural and phenotyping applications [1]. Despite significant progress, these models tend to perform best when trained on specific crop types rather than diverse datasets containing multiple crops. Research indicates that crop-specific models for RGB to NIR translation can outperform general-purpose models trained across all crop types. However, applying the correct model to each input image requires an intelligent selection mechanism [2].
To address this, the proposed work focuses on developing an optimized image classification algorithm capable of identifying crop types such as canola, wheat, and dry bean from aerial RGB imagery. By accurately classifying the crop type, the system can automatically route each image to its corresponding NIR generation model. This approach ensures high-quality NIR outputs while eliminating the need for manual sorting. The classifier must be both accurate and computationally efficient, as it serves as a guiding component within a larger end-to-end image generation pipeline without introducing significant processing overhead.
- Aslahishahri, Masoomeh, et al. "From RGB to NIR: Predicting of near infrared reflectance from visible spectrum aerial images of crops." Proceedings of the IEEE/CVF international conference on computer vision. 2021.
- Hossain, Md Yearat, et al. "Adaptive and automatic aerial image restoration pipeline leveraging pre-trained image restorer with lightweight Fully Convolutional Network." Expert Systems with Applications 259 (2025): 125210.
Relevant Majors: Computer Science, Electrical Engineering, Environmental Engineering
Assessing protein mislocalisation in the endomembrane system of mammalian cells
Prof. Jeremy C. Simpson
Functional analysis and annotation projects of the human genome have revealed that its encoded proteins localise to a vast array of compartments and structures throughout the cell. What is emerging is that single point mutations in individual genes can result in dramatic changes to the subcellular distribution of the encoded protein, which in turn can lead to disease. Recently, one large-scale study has attempted to catalogue this phenomenon for more than 1,200 human genes (1), making it the most comprehensive and systematic study to date examining protein mislocalisation. One striking observation in this work is the large number of proteins associated with compartments of the endomembrane system that show mislocalisation (2). Importantly, the full extent of the effects of this mislocalisation of many of these proteins in the wider context of cell health and function remain unknown.
This project will involve the molecular cloning of several of these identified genes, followed by determination of their subcellular localisation in cultured mammalian cells using advanced fluorescence microscopy methods. Key techniques will include bioinformatic and network analysis tools, molecular (gene) cloning, GFP-tagging, mammalian cell culture and fluorescence microscopy.
More details of the work of the lab can be found at: www.ucd.ie/hcs
- Lacoste et al. (2024) Cell 187(23):6725-6741.
- Dornan & Simpson (2024) Cell Genomics 4:100695.
Relevant Majors: Biomedical Sciences
What Can Collagen’s Piezoelectricity Reveal About Health and Aging?
Supervisor: Dr Brian Rodriguez, Dr Qiancheng Zhang
Piezoelectric materials are all around you, in microphones, lighters, watches, and ultrasound transducers, but they are also in you! Collagen is the most abundant protein in the human body, and it is piezoelectric - it generates an electric charge under mechanical stress. Studying collagen piezoelectricity can lead to biomedical applications and provide a deeper insight into biological processes, explaining for instance why astronauts lose bone density.
Structural differences in collagen, such as those observed in conditions like osteogenesis imperfecta (where collagen contains three alpha-1 chains instead of the typical two alpha-1 and one alpha-2 chains found in healthy collagen), significantly influence its mechanical and electromechanical properties. This project involves characterizing the nanoscale piezoresponse of collagen and comparing it across samples sourced from healthy, unhealthy, and aging tissues.
As part of the Nanoscale Function Group at University College Dublin (https://www.nanofunction.org), you will work within a multidisciplinary team exploring the intersection of physics, materials science, and biomedicine. Students will learn Atomic Force Microscopy, specialised electrical characterization modes, and sample preparation workflows. You will collect, interpret, and present electromechanical data, evaluating how collagen structure influences its piezoelectric performance. This opportunity provides hands-on experience with advanced techniques and builds invaluable skills in nanotechnology and biomedicine.
Relevant Majors: Physical sciences, life/health sciences, biomedical sciences, interdisciplinary sciences, engineering
DIY Atomic Force Microscope for Outreach and Education
Supervisor: Dr Brian Rodriguez, Dr Qiancheng Zhang
Atomic force microscopy (AFM) is one of the most powerful tools for imaging and manipulating matter at the nanoscale, yet its principles can be difficult to visualize without hands-on experience. This project focuses on the optimisation of DIY and LEGO AFM systems that demonstrate nanoscale sensing concepts and the development of engaging and accessible outreach activities for schools, science festivals, and the wider public.
Students will learn the core concepts of scanning probe microscopy and gain experience on research AFM systems. Students can apply AFM to materials that interest them and create content around their investigations.
Working with the Nanoscale Function Group at University College Dublin (https://www.nanofunction.org), you will join a highly collaborative environment engaged in research, education, and public engagement. This project is ideal for students who enjoy building things and communicating complex scientific ideas in accessible ways.
Relevant Majors: Physical sciences, engineering, computer science, education/outreach, interdisciplinary sciences, chemistry, life/health sciences, biomedical sciences, science communication
Fibrillation of Diabetes Peptide Drugs: Understanding Insulin and Glucagon Stability
Supervisor: Dr Brian Rodriguez, Dr Qiancheng Zhang
Therapeutic peptides such as insulin and glucagon are essential medicines used by millions. Their activity and stability are influenced by their molecular structure and formulation - fast-acting and long-acting insulin analogues have different stabilizers and structural modifications. These peptides can fibrillate into amyloids, similar to those associated with neurodegenerative diseases, but their formulations are designed to prevent this from happening, e.g., glucagon is provided dry and needs to be mixed before injection.
This project investigates how glucagon, fast-acting insulin, and slow-acting insulin fibrillate under controlled laboratory conditions. Students will vary parameters such as pH, agitation, temperature, and concentration to induce or suppress fibril formation, using atomic force microscopy (AFM) to quantify fibrillation parameters. The goal is to understand how formulation choices translate to real-world handling, stability, and clinical use and to translate the findings into an engaging patient and public-facing explainer. Time-allowing, we will explore the influence of nanoplastics on amyloid peptide fibrillation, linking peptide aggregation pathways with environmental nanosafety.
Working with the Nanoscale Function Group at University College Dublin (https://www.nanofunction.org), you will join a highly collaborative environment engaged in research, education, and public engagement. This project provides experience in biophysics, spectroscopy, microscopy, experimental design, and public engagement.
Relevant Majors: Physical sciences, chemistry, life/health sciences, biomedical sciences, pharmacology, environmental science, interdisciplinary sciences, science communication, engineering
The following information is vetted and provided by the American Association of Collegiate Registrars and Admissions Officers (AACRAO) on the Electronic Database for Global Education (EDGE).
| Letter Grade | Percentage | Ranking | U.S. Equivalent |
| A+/A/A- | 70 - 100% | First Class Honours | A |
| B+/B/B- | 60 - 69% | Second Class Honours Upper | B+ |
| C+/C/C- | 50 - 59% | Second Class Honours Lower | B |
| D+ | 45 - 49% | Third Class Honours | C+ |
| D/D- | 40 - 44% | Pass | C |
| F | 0 - 39% | Fail | F |